Difference between revisions of "iRefScape 1.0 Installation"
| (21 intermediate revisions by 4 users not shown) | |||
| Line 32: | Line 32: | ||
# Restart Cytoscape. | # Restart Cytoscape. | ||
# Select iRefIndexx.x from Cytoscape's "Plugins" menu. | # Select iRefIndexx.x from Cytoscape's "Plugins" menu. | ||
| − | # When the plugin is started for the first time, it will download the | + | # When the plugin is started for the first time, it will download the dataset. |
| − | # You will be prompted to select an installation directory for the data and indices. The recommended place would be the plugin home directory which | + | # You will be prompted to select an installation directory for the data and indices. The recommended place would be the plugin home directory which will appear as the recommended directory. The data and indices require a minimum disk space of 1 GB at this location. Using this default directory is strongly encouraged. |
# If the default directory is selected and is not accessible, you will be prompted to restart Cytoscape. If this message appears on second or subsequent attempts, this may mean that there is a problem accessing this directory. To avoid such problems, ensure that you have write access to the directory and at least 1GB of free space on the disk. | # If the default directory is selected and is not accessible, you will be prompted to restart Cytoscape. If this message appears on second or subsequent attempts, this may mean that there is a problem accessing this directory. To avoid such problems, ensure that you have write access to the directory and at least 1GB of free space on the disk. | ||
| − | # The downloading and installation of indices usually takes less than 5 minutes if you have a internet connection of 10Mb/sec or more. However | + | # The downloading and installation of indices usually takes less than 5 minutes if you have a internet connection of 10Mb/sec or more. However installation may take between 15 and 30 minutes depending on your internet connection. |
# The installation process should now complete itself. | # The installation process should now complete itself. | ||
# If the installation was successful, you will see a new iRefScape panel near the "Network" tab of Cytoscape as well as an iRefScape menu (blue) at the top of the Cytoscape interface. Also, you will see an Information frame with several tabs in it ("Messages", "Search history", "Query helper", and so on). This window can be closed. | # If the installation was successful, you will see a new iRefScape panel near the "Network" tab of Cytoscape as well as an iRefScape menu (blue) at the top of the Cytoscape interface. Also, you will see an Information frame with several tabs in it ("Messages", "Search history", "Query helper", and so on). This window can be closed. | ||
<br clear="all" /> | <br clear="all" /> | ||
| − | |||
| − | |||
| − | |||
| − | |||
=== Using the iRefIndex Installation site === | === Using the iRefIndex Installation site === | ||
| Line 58: | Line 54: | ||
# Click "Add" and for each of the following fields, enter the suggested values: | # Click "Add" and for each of the following fields, enter the suggested values: | ||
#* Name: <tt>iRefScape</tt> | #* Name: <tt>iRefScape</tt> | ||
| − | #* URL: <tt> | + | #* URL: <tt>http://irefindex.org/download/irefindex/Cytoscape/plugin/current/plugins_irefindex.xml</tt> |
| Line 69: | Line 65: | ||
# Go to the "Completing the installation" step above. | # Go to the "Completing the installation" step above. | ||
| − | === Manual Installation === | + | === Install from file === |
| + | |||
| + | If the plugin download manager fails to initialise or if you encounter problems using it, the plugin can be manually downloaded from the following locations: | ||
| + | |||
| + | * The Cytoscape plugin repository at http://chianti.ucsd.edu/cyto_web/plugins/ (look for iRefScape under "Network and Attribute I/O"). Locate the link "Download Jar/zip: click here" and download the plugin to a easily locatable place (such as the desktop). | ||
| + | * Directly from the Cytoscape site at http://chianti.ucsd.edu/cyto_web/plugins/displayplugininfo.php?name=iRefScape (the latest version is listed last). Locate the link "Download Jar/zip: click here" and download the plugin to a easily locatable place (such as the desktop). | ||
| + | *From the iRefIndex download site: http://irefindex.org/download/irefindex/Cytoscape/plugin/current/iRefScape.jar | ||
| + | |||
| + | After downloading the plugin can be installed by following the steps below: | ||
| + | |||
| + | # Start Cytoscape. | ||
| + | # Make sure that no other versions of the iRefScape plugin are already installed. If you see iRefScape, uninstall the plugin and restart Cytoscape before proceeding. | ||
| + | # From the Cytoscape menu select "Plugins" -> "Install plugin from file". | ||
| + | # When the file open dialogue window appears, select the downloaded file (<tt>iRefScape.jar</tt>). | ||
| + | # After installation is complete, restart Cytoscape. Do not attempt to activate the iRefScape plugin before restarting Cytoscape. | ||
| + | # After restarting, activate the plugin by selecting iRefScape from the Cytoscape "Plugin" menu and follow instructions to download the dataset. | ||
| + | |||
| + | {{Note| | ||
| + | Please note that this method does not solve firewall-related issues. | ||
| + | }} | ||
| + | |||
| + | === Manual Installation (if you have problems installing with the Cytoscape plugin manager) === | ||
| + | |||
| + | If you are still having problems, you can try manually downloading and installing the files. This is especially useful if you are having firewall problems while getting the plugin or the data. | ||
| + | |||
| + | Before starting: | ||
| + | |||
| + | <ol> | ||
| + | <li>Make sure that Cytoscape is not running.</li> | ||
| + | <li>Remove any other installations of iRefScape (or iRefIndex) or directories created by those installations. These directories look like the following: | ||
| + | <pre>.../Cytoscape_vx.x.x/plugins/iRefScape</pre></li> | ||
| + | <li>Go to http://irefindex.org/download/irefindex/Cytoscape/plugin/current/</li> | ||
| + | <li>Download <tt>iRefScape.jar</tt>.</li> | ||
| + | <li>Download <tt>iRefDATA_mmddyyyy.irfz</tt> (where <tt>mmddyyyy</tt> is the date stamp for the file).</li> | ||
| + | <li>Copy the <tt>iRefScape.jar</tt> file to the <tt>plugins</tt> directory of your Cytoscape installation. It appears necessary to use the <tt>plugins</tt> directory within the Cytoscape software's own directory structure rather than, for example, the one found within the <tt>.cytoscape</tt> directory hierarchy in your home directory (which is where the automatic installation tends to install the software).</li> | ||
| + | <li>Copy the <tt>iRefDATA_mmddyyyy.irfz</tt> file to the same directory or anywhere else where you have permissions to write files.</li> | ||
| + | <li>Start Cytoscape.</li> | ||
| + | <li>Select iRefIndex from the Cytoscape "Plugins" menu.</li> | ||
| + | <li>When prompted for an installation directory, select the directory where you placed the <tt>iRefDATA_mmddyyyy.irfz</tt> file.</li> | ||
| + | <li>The plugin will complete the installation without having to first download the <tt>iRefDATA_mmddyyyy.irfz</tt> file.</li> | ||
| + | </ol> | ||
| + | |||
| + | === Uninstalling the Plugin=== | ||
| + | |||
| + | # From the Cytoscape Plugins menu, select "Currently Installed" -> "Network and Attribute I/O Plugins" and choose the iRefScape plugin. | ||
| + | # Then, click the "Delete" button and confirm. | ||
| + | # Click "Close" to leave the "Manage Plugins" dialogue and restart Cytoscape. | ||
| + | # The plugin will be deleted when you restart Cytoscape. | ||
| + | |||
| − | + | === Increasing memory available to Cytoscape === | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | < | + | If the Cytoscape interface becomes unresponsive (especially when working with large graphs), take a look at the [http://cytoscape.wodaklab.org/wiki/How_to_increase_memory_for_Cytoscape How to increase memory for Cytoscape] page to increase the memory made available to Cytoscape. |
| + | |||
| + | == Troubleshooting Platform-Specific Issues == | ||
| + | |||
| + | === Windows Vista and Windows XP === | ||
| + | |||
| + | You will require write privileges to the installation directory not only during installation but also during operation of the plugin. If the plugin disappears from the "Plugins" menu or it does not appear at all, then Cytoscape has to be started as an administrator. This could be done by right-clicking on the Cytoscape shortcut and selecting "Run as" option. When requesting help for such situations, please include a copy-paste of the Cytoscape error console ("Help" -> "Error console"). (To copy-paste, open the error console, click inside, press <tt>Ctrl-A</tt> (to select all) then <tt>Ctrl-C</tt> (to copy). The selection can then be pasted into a message using <tt>Ctrl-V</tt>.) | ||
| + | |||
| + | === Mac OS X === | ||
| + | |||
| + | * Please verify that Java 1.6 or later is available. ''The plugin will not work unless Java 1.5 or later is installed.'' | ||
| + | * Increase the <tt>Xmx</tt> setting to <tt>512m</tt> and the <tt>Xms</tt> setting to <tt>512m</tt> in <tt>cytoscape.sh</tt>. | ||
| + | * This version of the plugin is not extensively tested on Mac OS and we are sorry that the support we could provide for Mac users is limited at this point. | ||
| + | |||
| + | === Unix/Linux === | ||
| + | |||
| + | * Please verify that Java 1.6 or later is available. | ||
| + | ** The "Java SE Development Kit (JDK)" from http://java.sun.com/javase/downloads/index.jsp is suitable. The Sun/Oracle version of Java may be available as a package for your distribution. For example, the <tt>sun-java6-bin</tt> package for Ubuntu provides a compatible version of Java. | ||
| + | ** The OpenJDK packages from http://openjdk.java.net/install/ may also be suitable and are often available via distributions. For example, OpenJDK 6 for [http://packages.debian.org/search?keywords=openjdk-6-jdk&searchon=names&suite=all§ion=all Debian] and [http://packages.ubuntu.com/search?keywords=openjdk-6-jdk&searchon=names&suite=all§ion=all Ubuntu]. | ||
| + | ** After installing a new version of Java for Debian or Ubuntu, you may need to use the <tt>update-alternatives</tt> command to indicate that you would like to use this new version instead of any existing version, or you may need to add the precise location of the new <tt>java</tt> program to your <tt>PATH</tt>. | ||
| + | * You will require write privileges to the installation directory not only during installation but also during operation. | ||
| + | * The plugin behavior is proven to be very stable in Unix/Linux environments. | ||
| + | * To improve font appearance, try adding the following environment variable definition to your configuration (in <tt>.bashrc</tt> or <tt>.bash_profile</tt>), as described in this [http://wiki.archlinux.org/index.php/Java_Fonts_-_Sun_JRE guide to Java fonts for Arch Linux]: | ||
| + | |||
| + | export _JAVA_OPTIONS='-Dawt.useSystemAAFontSettings=lcd' | ||
| − | == Troubleshooting | + | == Troubleshooting Security Issues== |
The plugin has two components: | The plugin has two components: | ||
| Line 93: | Line 154: | ||
If the plugin manager is used for the installation (as of Cytoscape 2.6.3 and later), the plugin will be placed in a directory under the home area and a record file is kept in a file named <tt>track_plugins.xml</tt>. The location of these would as follows: | If the plugin manager is used for the installation (as of Cytoscape 2.6.3 and later), the plugin will be placed in a directory under the home area and a record file is kept in a file named <tt>track_plugins.xml</tt>. The location of these would as follows: | ||
| − | * Linux: <tt>~/.cytoscape/2. | + | * Linux: <tt>~/.cytoscape/2.8</tt> |
| − | * Windows: <tt>''<user's home directory>''\.cytoscape\2. | + | * Windows: <tt>''<user's home directory>''\.cytoscape\2.8</tt> |
The entry corresponding to iRefScape plugin would looks like follows: | The entry corresponding to iRefScape plugin would looks like follows: | ||
| Line 143: | Line 204: | ||
<br clear="all" /> | <br clear="all" /> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
== About the Installation Files == | == About the Installation Files == | ||
| − | During installation the plugin downloads protein-protein interaction data from iRefIndex ( | + | During installation the plugin downloads protein-protein interaction data from iRefIndex (http://irefindex.org/download/irefindex/Cytoscape/plugin/current/) in a compressed form. The downloaded files will reside in the <tt>iRefIndex</tt> directory under the Cytoscape plugin folder. It is recommended that the files in this directory '''not''' be modified as this may lead to unpredictable results. In particular, please do not open them in word-processing software (such as Microsoft Office). After successful installation you will also find some index files and serialized Java object files which will be used in the search and load operations. |
=== Brief Description of the Files === | === Brief Description of the Files === | ||
Latest revision as of 20:08, 14 March 2013
Contents
- 1 Installation
- 1.1 Before You Begin
- 1.2 Using the Cytoscape Plugin menu
- 1.3 Completing the Installation
- 1.4 Using the iRefIndex Installation site
- 1.5 Install from file
- 1.6 Manual Installation (if you have problems installing with the Cytoscape plugin manager)
- 1.7 Uninstalling the Plugin
- 1.8 Increasing memory available to Cytoscape
- 2 Troubleshooting Platform-Specific Issues
- 3 Troubleshooting Security Issues
- 4 About the Installation Files
Installation
Before You Begin
- These instructions assume that you have downloaded and installed the Cytoscape application. See http://cytoscape.org/ for a manual and a set of tutorials which describe the installation and use of Cytoscape.
- Make sure you have an active Internet connection. During installation the plugin will download files from the iRefIndex FTP site.
- Make sure you have latest Java environment installed: Java 1.6 or later is required.
- If you have a plugin called iRefIndex installed, it is recommended that you first delete it, selecting the "Manage Plugins" option in the "Plugins" menu. This will only be the case for people who may have tried earlier development versions of the plugin.
- Start Cytoscape. You must start Cytoscape as a user that has write privileges to the target directory.
- From the menu select "Plugins" -> "Manage Plugins".
- Select "Manage plugins" from the plugin menu.
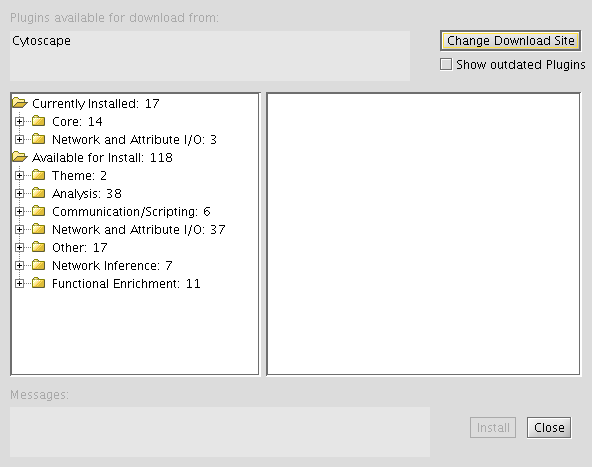
- Locate "Available for Install" then "Network and Attribute I/O", verifying that iRefScape is listed in this category.
- Select the iRefScape plugin and then click "Install" (in the bottom-right-hand corner of the window).
- When you see the message "iRefScape v.x.x install complete" click "Close" to leave the "Manage Plugins" dialogue.
- Close Cytoscape.
- Go to the next section to complete the setup.
Completing the Installation
- Restart Cytoscape.
- Select iRefIndexx.x from Cytoscape's "Plugins" menu.
- When the plugin is started for the first time, it will download the dataset.
- You will be prompted to select an installation directory for the data and indices. The recommended place would be the plugin home directory which will appear as the recommended directory. The data and indices require a minimum disk space of 1 GB at this location. Using this default directory is strongly encouraged.
- If the default directory is selected and is not accessible, you will be prompted to restart Cytoscape. If this message appears on second or subsequent attempts, this may mean that there is a problem accessing this directory. To avoid such problems, ensure that you have write access to the directory and at least 1GB of free space on the disk.
- The downloading and installation of indices usually takes less than 5 minutes if you have a internet connection of 10Mb/sec or more. However installation may take between 15 and 30 minutes depending on your internet connection.
- The installation process should now complete itself.
- If the installation was successful, you will see a new iRefScape panel near the "Network" tab of Cytoscape as well as an iRefScape menu (blue) at the top of the Cytoscape interface. Also, you will see an Information frame with several tabs in it ("Messages", "Search history", "Query helper", and so on). This window can be closed.
Using the iRefIndex Installation site
In rare cases, the Cytoscape plugin site may be down. In this case, you have the option of using the iRefScape plugin site.
- Start Cytoscape. You must start Cytoscape as a user that has write privileges to the target directory.
- From the menu select "Plugins" -> "Manage Plugins".
To use the iRefScape plugin site:
- Click on "Settings" tab.
- Click "Add" and for each of the following fields, enter the suggested values:
To install the plugin:
- Locate "Available for Install" then "Network and Attribute I/O", verifying that iRefScape is listed in this category.
- Select the iRefScape plugin and then click "Install" (in the bottom-right-hand corner of the window).
- Click "Close" to leave the "Manage Plugins" dialogue.
- Restart Cytoscape.
- Go to the "Completing the installation" step above.
Install from file
If the plugin download manager fails to initialise or if you encounter problems using it, the plugin can be manually downloaded from the following locations:
- The Cytoscape plugin repository at http://chianti.ucsd.edu/cyto_web/plugins/ (look for iRefScape under "Network and Attribute I/O"). Locate the link "Download Jar/zip: click here" and download the plugin to a easily locatable place (such as the desktop).
- Directly from the Cytoscape site at http://chianti.ucsd.edu/cyto_web/plugins/displayplugininfo.php?name=iRefScape (the latest version is listed last). Locate the link "Download Jar/zip: click here" and download the plugin to a easily locatable place (such as the desktop).
- From the iRefIndex download site: http://irefindex.org/download/irefindex/Cytoscape/plugin/current/iRefScape.jar
After downloading the plugin can be installed by following the steps below:
- Start Cytoscape.
- Make sure that no other versions of the iRefScape plugin are already installed. If you see iRefScape, uninstall the plugin and restart Cytoscape before proceeding.
- From the Cytoscape menu select "Plugins" -> "Install plugin from file".
- When the file open dialogue window appears, select the downloaded file (iRefScape.jar).
- After installation is complete, restart Cytoscape. Do not attempt to activate the iRefScape plugin before restarting Cytoscape.
- After restarting, activate the plugin by selecting iRefScape from the Cytoscape "Plugin" menu and follow instructions to download the dataset.
|
Please note that this method does not solve firewall-related issues. |
Manual Installation (if you have problems installing with the Cytoscape plugin manager)
If you are still having problems, you can try manually downloading and installing the files. This is especially useful if you are having firewall problems while getting the plugin or the data.
Before starting:
- Make sure that Cytoscape is not running.
- Remove any other installations of iRefScape (or iRefIndex) or directories created by those installations. These directories look like the following:
.../Cytoscape_vx.x.x/plugins/iRefScape
- Go to http://irefindex.org/download/irefindex/Cytoscape/plugin/current/
- Download iRefScape.jar.
- Download iRefDATA_mmddyyyy.irfz (where mmddyyyy is the date stamp for the file).
- Copy the iRefScape.jar file to the plugins directory of your Cytoscape installation. It appears necessary to use the plugins directory within the Cytoscape software's own directory structure rather than, for example, the one found within the .cytoscape directory hierarchy in your home directory (which is where the automatic installation tends to install the software).
- Copy the iRefDATA_mmddyyyy.irfz file to the same directory or anywhere else where you have permissions to write files.
- Start Cytoscape.
- Select iRefIndex from the Cytoscape "Plugins" menu.
- When prompted for an installation directory, select the directory where you placed the iRefDATA_mmddyyyy.irfz file.
- The plugin will complete the installation without having to first download the iRefDATA_mmddyyyy.irfz file.
Uninstalling the Plugin
- From the Cytoscape Plugins menu, select "Currently Installed" -> "Network and Attribute I/O Plugins" and choose the iRefScape plugin.
- Then, click the "Delete" button and confirm.
- Click "Close" to leave the "Manage Plugins" dialogue and restart Cytoscape.
- The plugin will be deleted when you restart Cytoscape.
Increasing memory available to Cytoscape
If the Cytoscape interface becomes unresponsive (especially when working with large graphs), take a look at the How to increase memory for Cytoscape page to increase the memory made available to Cytoscape.
Troubleshooting Platform-Specific Issues
Windows Vista and Windows XP
You will require write privileges to the installation directory not only during installation but also during operation of the plugin. If the plugin disappears from the "Plugins" menu or it does not appear at all, then Cytoscape has to be started as an administrator. This could be done by right-clicking on the Cytoscape shortcut and selecting "Run as" option. When requesting help for such situations, please include a copy-paste of the Cytoscape error console ("Help" -> "Error console"). (To copy-paste, open the error console, click inside, press Ctrl-A (to select all) then Ctrl-C (to copy). The selection can then be pasted into a message using Ctrl-V.)
Mac OS X
- Please verify that Java 1.6 or later is available. The plugin will not work unless Java 1.5 or later is installed.
- Increase the Xmx setting to 512m and the Xms setting to 512m in cytoscape.sh.
- This version of the plugin is not extensively tested on Mac OS and we are sorry that the support we could provide for Mac users is limited at this point.
Unix/Linux
- Please verify that Java 1.6 or later is available.
- The "Java SE Development Kit (JDK)" from http://java.sun.com/javase/downloads/index.jsp is suitable. The Sun/Oracle version of Java may be available as a package for your distribution. For example, the sun-java6-bin package for Ubuntu provides a compatible version of Java.
- The OpenJDK packages from http://openjdk.java.net/install/ may also be suitable and are often available via distributions. For example, OpenJDK 6 for Debian and Ubuntu.
- After installing a new version of Java for Debian or Ubuntu, you may need to use the update-alternatives command to indicate that you would like to use this new version instead of any existing version, or you may need to add the precise location of the new java program to your PATH.
- You will require write privileges to the installation directory not only during installation but also during operation.
- The plugin behavior is proven to be very stable in Unix/Linux environments.
- To improve font appearance, try adding the following environment variable definition to your configuration (in .bashrc or .bash_profile), as described in this guide to Java fonts for Arch Linux:
export _JAVA_OPTIONS='-Dawt.useSystemAAFontSettings=lcd'
Troubleshooting Security Issues
The plugin has two components:
- The Java executable file (iRefScape_x.x.jar) which is the software component providing the plugin functionality.
- The data component containing protein-protein interaction data.
Both these components are required for the functioning of the plugin. The installation section above explains how to install the executable components, and once this component is installed and the plugin is loaded, the data component will then be downloaded by the plugin itself. The downloaded file is compressed and has the extension .irfz and a size of around 200 MB. After the download is complete this file will be partially uncompressed (the indices and few other files required for immediate operation will be extracted from the complex file). However, the bulk of the data will remain compressed and will only be uncompressed when needed. Thus, the speed of query execution will increase with usage and the size of the iRefIndex directory will increase. Therefore, although 250MB of available free space is enough during installation, the plugin requires up to 1GB of space for its operation.
If the plugin manager is used for the installation (as of Cytoscape 2.6.3 and later), the plugin will be placed in a directory under the home area and a record file is kept in a file named track_plugins.xml. The location of these would as follows:
- Linux: ~/.cytoscape/2.8
- Windows: <user's home directory>\.cytoscape\2.8
The entry corresponding to iRefScape plugin would looks like follows:
<plugin> <uniqueID>999</uniqueID> <name>iRefScape</name> <description>.<br>http://irefindex.uio.no/wiki/README_Cytoscape_plugin_0.8x<p></description> <cytoscapeVersion>2.6</cytoscapeVersion> <url>ftp://ftp.no.embnet.org/irefindex/Cytoscape/plugin/archive/beta8/iRefScape_0_83.jar</url> <downloadUrl>ftp://ftp.no.embnet.org/irefindex/Cytoscape/plugin/current/plugins_irefindex.xml</downloadUrl> <category>Network and Attribute I/O</category> <releaseDate>MAY 20, 2010</releaseDate> <pluginVersion>0.83</pluginVersion> <classname>cytoscape.no.uio.biotek.Main</classname> <projectUrl>http://irefindex.uio.no</projectUrl> <filetype>jar</filetype> <installLocation>.../.cytoscape/2.6/plugins/iRefScape-0.83</installLocation> <license> <text /> </license> <filelist> <file>.../.cytoscape/2.6/plugins/iRefScape-0.83/iRefScape.jar</file> </filelist> <authorlist> <author> <name>Ian Donaldson and Sabry Razick</name> <institution>Biotechnology Centre of Oslo, University of Oslo</institution> </author> </authorlist> </plugin>
Note: If you encounter an exception during Cytoscape's start-up process due to a plugin failure and there is no way of uninstalling it, you could remove the entry for the corresponding plugin from this file when Cytoscape is not running. First, take a backup of the file, then delete the plugin entry resembling that shown above for the plugin concerned. Finally, save the file and restart Cytoscape.
The download and installation of the plugin executable is managed by the core functionality provided by Cytoscape, and if the plugin complies with the requirements defined for the plugin framework, the installation should proceed smoothly. However, some users have encountered problems when installing the data component; this is mainly due to privilege and security issues.
- Privilege issues arise when a user does not have access to the place where the data component is to be installed. This directory should permit write access for that user.
- Security issues are mainly encountered with Microsoft Windows-based operating systems. When software tries to perform a suspicious operation, the operation is blocked and the user is prompted to confirm that this is something they have chosen to do themselves. If the user selects "block", the iRefScape plugin installation fails and it may not be possible to attempt to install the plugin again until the blocking action is revoked manually. Although antivirus products are also known to interfere with software installers, the plugin installation process has been tested on systems running several popular antivirus products, but has not revealed any issues relating to them at the time of writing. If you encounter any such issues, please report to these to the iRefIndex Google Group.
The behaviour of the plugin when installed using the plugin manager is different before and after Cytoscape is restarted. After the installation, the plugin will appear in the plugin manager and could be activated, however it is not aware of its environment: for example, it does not know its installation location. Therefore it is highly recommended that Cytoscape be restarted immediately after installing the plugin, and that the plugin is not activated before the first restart (do not click on the "iRefScape" menu entry). When a manual installation is performed, the plugin can be activated after starting Cytoscape for the first time, since Cytoscape is not running during the installation process.
Some of the reasons behind distributing the data with the plugin rather than more popular approaches such as the use of Web services include the following:
- Execution speed
- No need for a fast internet connection after installation
- Hosting restrictions applying to the iRefScape/iRefIndex download site
- Privacy for the user: there is no monitoring of the types of searches the user performs; thus, the user's activities remain private
About the Installation Files
During installation the plugin downloads protein-protein interaction data from iRefIndex (http://irefindex.org/download/irefindex/Cytoscape/plugin/current/) in a compressed form. The downloaded files will reside in the iRefIndex directory under the Cytoscape plugin folder. It is recommended that the files in this directory not be modified as this may lead to unpredictable results. In particular, please do not open them in word-processing software (such as Microsoft Office). After successful installation you will also find some index files and serialized Java object files which will be used in the search and load operations.
Brief Description of the Files
- irft files: this file contains a ROG (Redundant Object Group) to PARAMETER mapping. This is a sort of index that maps identifiers like accessions and names to ROGs. The ROG is an integer representation of the ROGID. However, ROG is not stable like ROGID and it may be different from one version to another. However, from beta4 onwards the ROGID to ROG mapping will be propagated for backwards compatibility for live proteins (proteins which are not removed from original source).
- Irfm files: These are index files containing information about partners of each interaction.
- ROGS Directory: Contains protein attributes (Warning! Please do not try to open this directory in a file browser; the computer may crash)
- RIGS Directory: Contains interaction attributes (Warning! Please do not try to open this directory in a file browser; the computer may crash)
- Irfj files : these files contain the interaction and object data in a compressed form. When requested for the first time the information will be uncompressed to the RIGS or ROGS directory.